 The classification of protein
structural information, especially using the overall structure of
the protein (the fold) as the basis for the classification, is an
exciting area of research in structural biology. We were
interested in the complementary question, could we develop a
classification scheme based on the primary structure (residue
based) that would allow us to understand on the molecular level
the intricacies of protein structure.
The classification of protein
structural information, especially using the overall structure of
the protein (the fold) as the basis for the classification, is an
exciting area of research in structural biology. We were
interested in the complementary question, could we develop a
classification scheme based on the primary structure (residue
based) that would allow us to understand on the molecular level
the intricacies of protein structure.
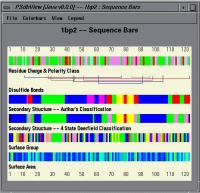
In this work, we present the Protein Structure Database (PSdb), a new protein database that relates secondary (e.g. Helix, Sheet, Turn, Random Coil), supersecondary (e.g., helix-helix interactions), and tertiary information (e.g. Solvent accessibility, internal relative distances, and ligand interactions) to the primary structure. The data for each protein is supplied on a residue by residue basis and encoded in a series of flat ASCII files.
Relationships between the various levels of structure (primary, secondary, tertiary) can be investigated visually using PSdbView, a graphical tool provided to view the information within the PSdb. This tool allows for side by side comparison of residue based data and includes a variety of standard mechanisms for visualizing protein data including Ramachandran plots, C(alpha)-C(alpha) distance plots, and differences in solvent accessible molecular surface area graphs (e.g., differences in the exposed surface with and without including either the ligands, metal ions or buried waters in the computations). PSdbView is written in Java, thus providing a platform independent means for exploring PSdb entries over the Internet.